Cophenetic metrics for phylogenetic trees, after Sokal and Rohlf
Correlation between metrics over phylogenetic trees with 60 leaves (Simulation)
Non-necessary binary phylogenetic trees
Spearman's correlation between cophenetic, nodal and RF metrics.
| | Cophenetic p=1 | Cophenetic p=2 | Nodalsp p=1 | Nodalsp p=2 | RF |
| Coph p=1 | 1.0 | 0.986886672255 | 0.732991855139 | 0.762110570162 | 0.107535731844 |
| Coph p=2 | | 1.0 | 0.783531973395 | 0.813692157103 | 0.119250141004 |
| Nsp p=1 | | | 1.0 | 0.989352552785 | 0.239181245199 |
| Nsp p=2 | | | | 1.0 | 0.218849067697 |
| RF | | | | | 1.0 |
2D histograms. A darker squeare means that there are more trees with that relationship.
| | Cophenetic p=2 | Nodalsp p=1 | Nodalsp p=2 | RF |
| Coph p=1 |  | 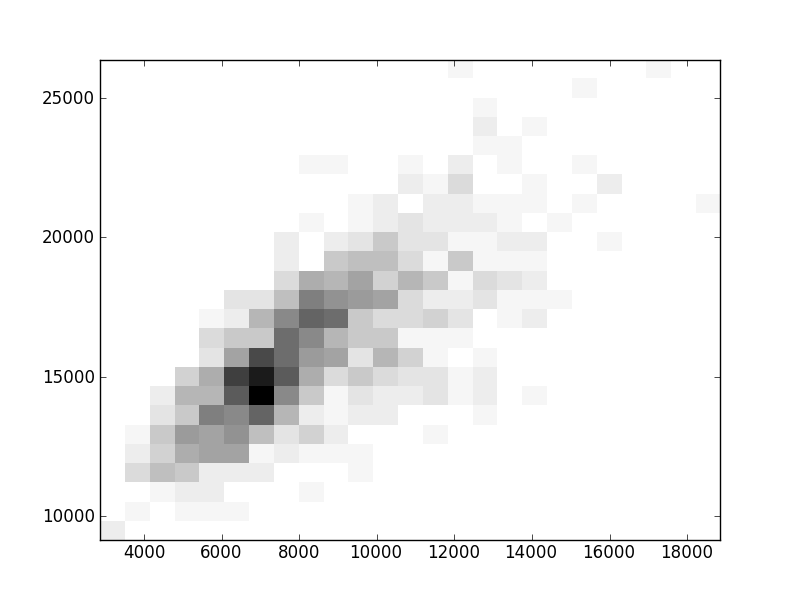 | 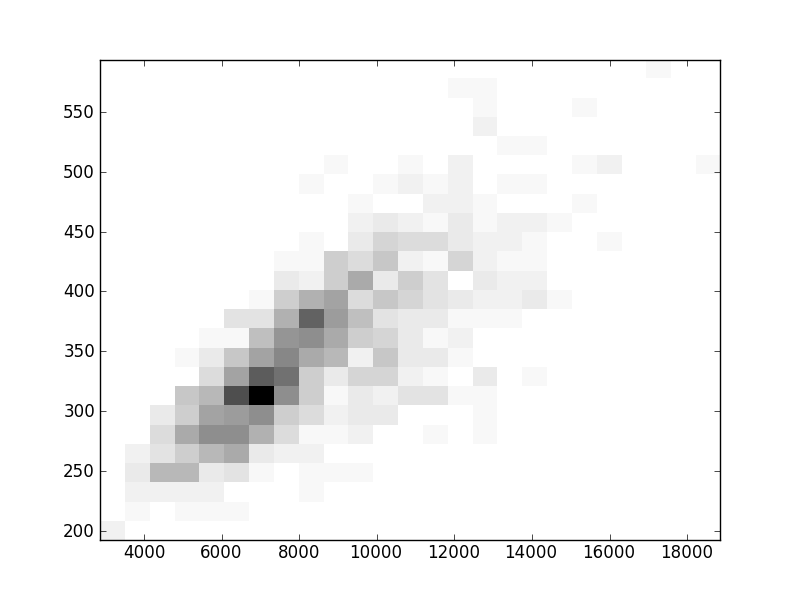 |  |
| Coph p=2 | | 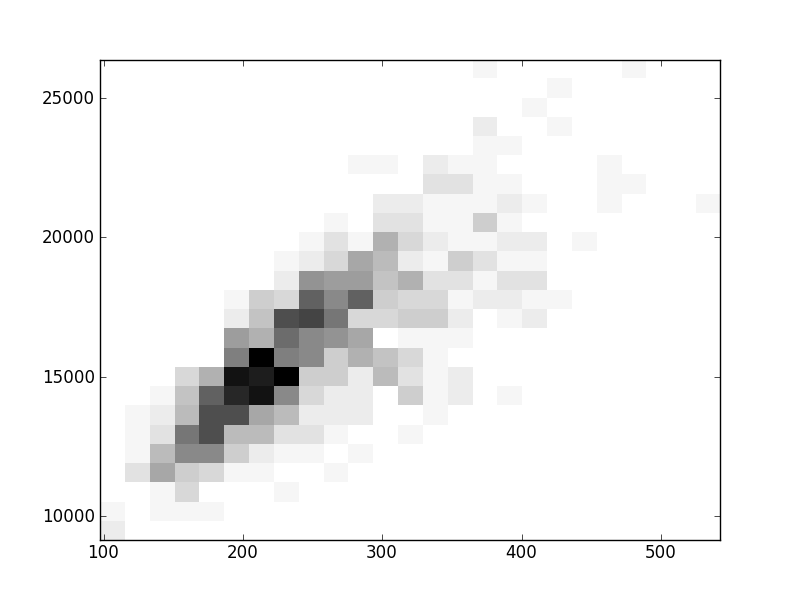 | 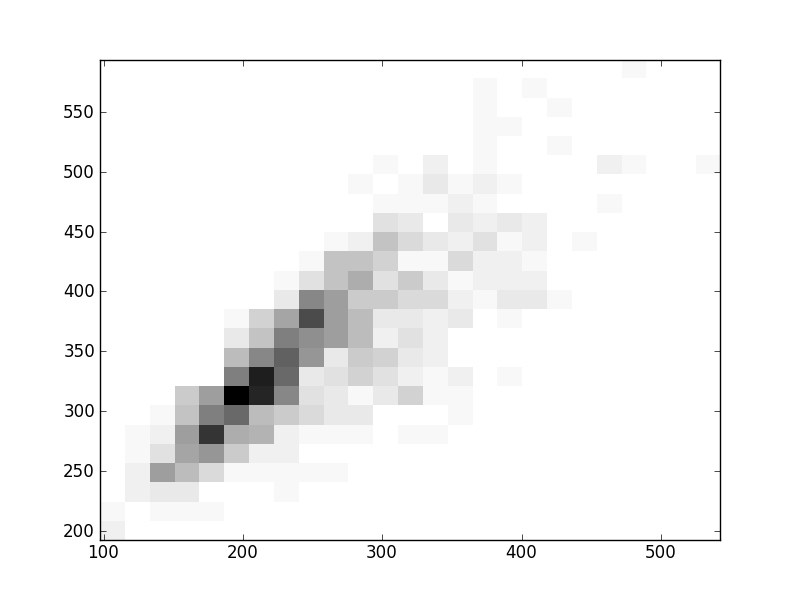 | 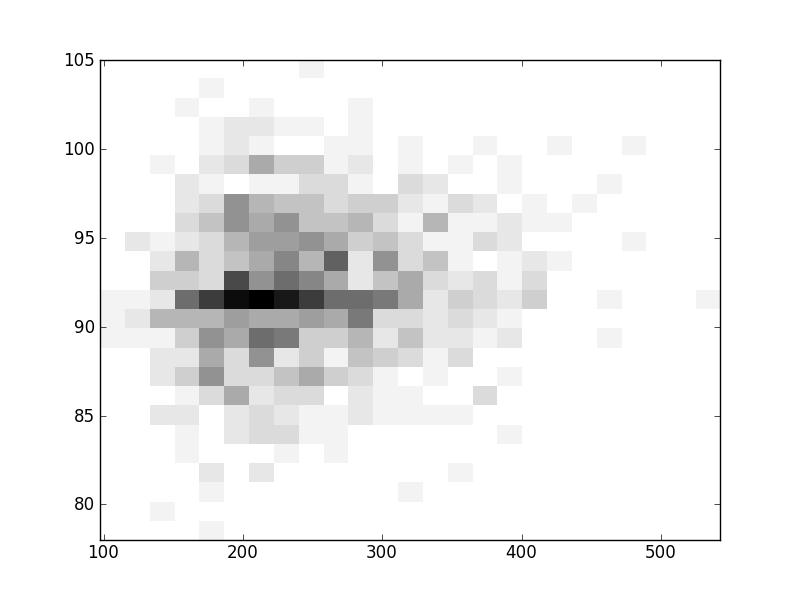 |
| Nsp p=1 | | |  | 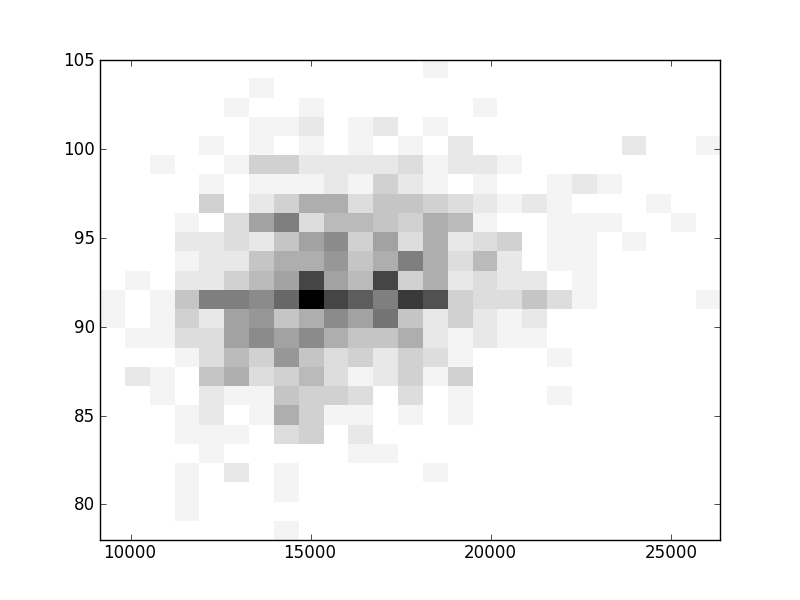 |
| Nsp p=2 | | | | 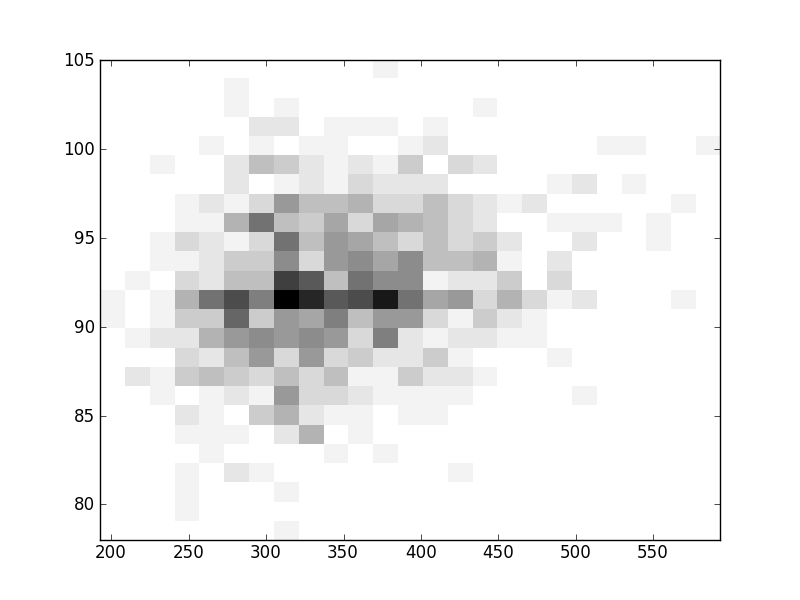 |
Binary phylogenetic trees
Spearman's correlation between cophenetic, nodal and RF metrics.
| | Cophenetic p=1 | Cophenetic p=2 | Nodalsp p=1 | Nodalsp p=2 | Nodalunsp p=1 | Nodalunsp p=2 | RF |
| Coph p=1 | 1.0 | 0.987363704233 | 0.75396883837 | 0.779191547568 | 0.401723406939 | 0.404946907239 | 0.0292204936767 |
| Coph p=2 | | 1.0 | 0.795401040162 | 0.822322208992 | 0.46785753735 | 0.471086892009 | 0.0284685349973 |
| Nsp p=1 | | | 1.0 | 0.988988054504 | 0.77358614221 | 0.775099770658 | 0.0337844360343 |
| Nsp p=2 | | | | 1.0 | 0.749178779381 | 0.752895960404 | 0.0293644965612 |
| Nunsp p=1 | | | | | 1.0 | 0.997626024447 | 0.0464444472901 |
| Nunsp p=2 | | | | | | 1.0 | 0.0431704425722 |
| RF | | | | | | | 1.0 |
2D histograms. A darker squeare means that there are more trees with that relationship.
| | Cophenetic p=2 | Nodalsp p=1 | Nodalsp p=2 | Nodalunsp p=1 | Nodalunsp p=2 | RF |
| Coph p=1 | 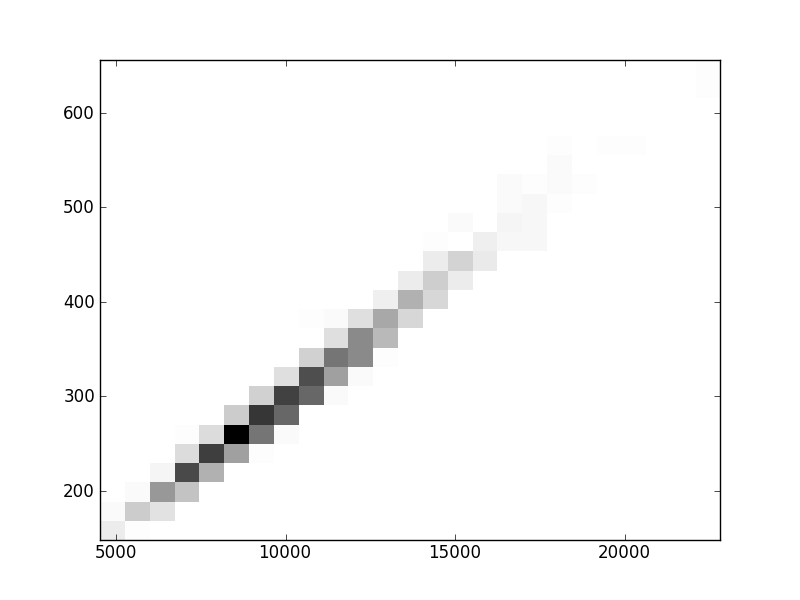 | 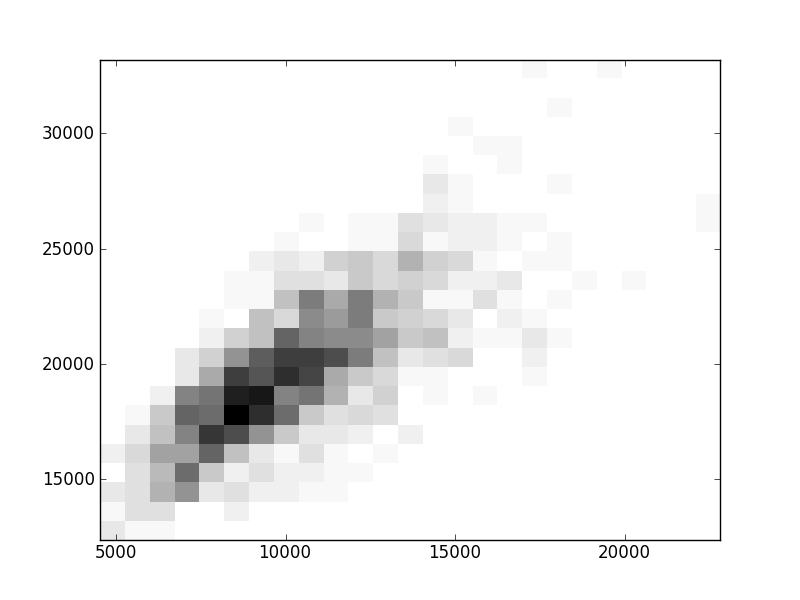 | 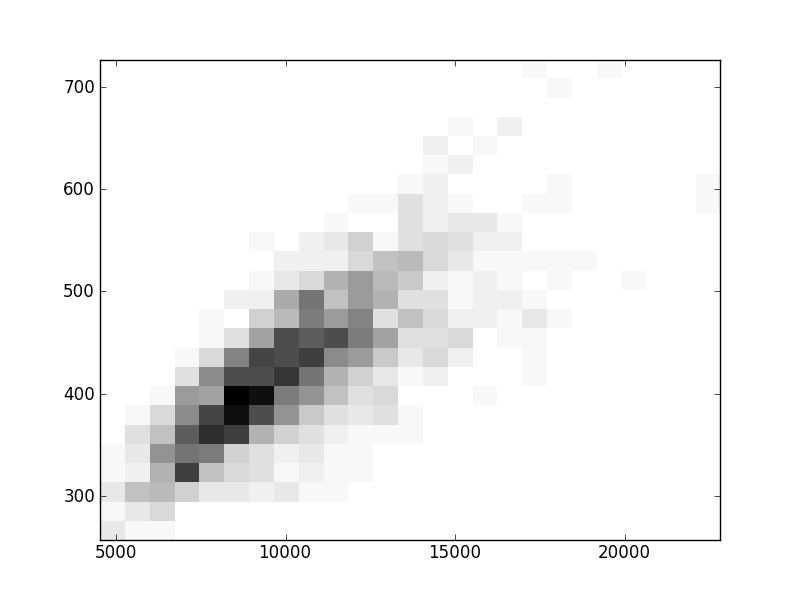 |  | 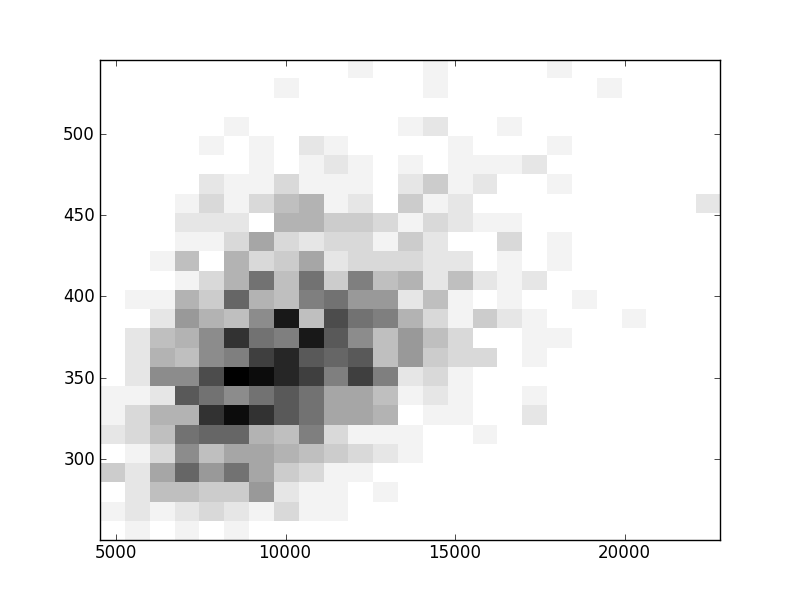 | 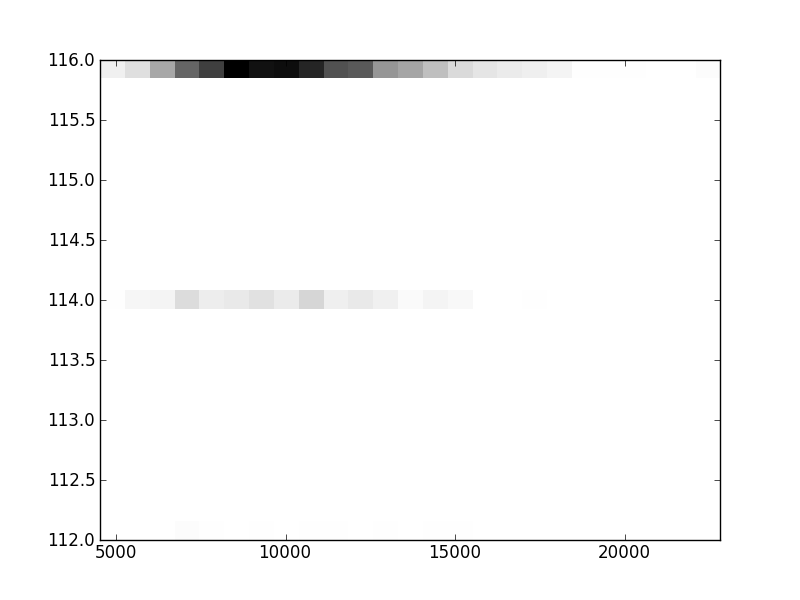 |
| Coph p=2 | | 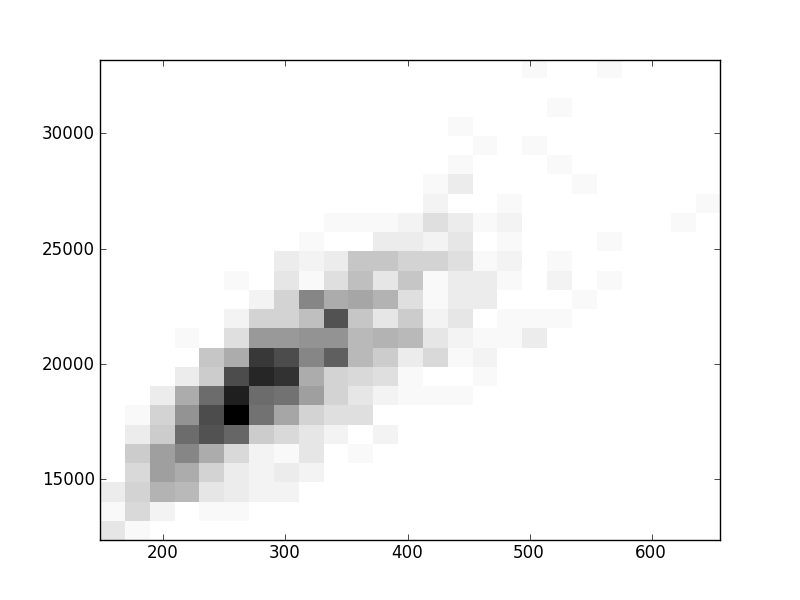 | 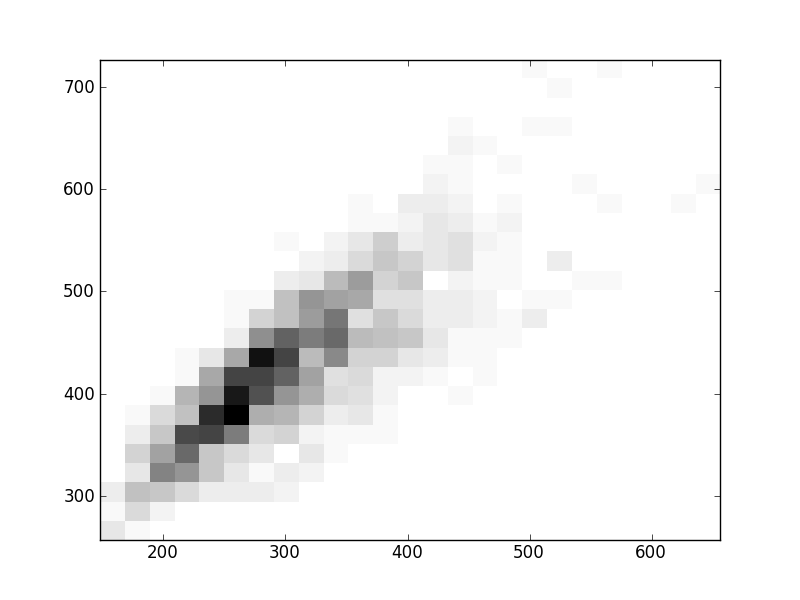 | 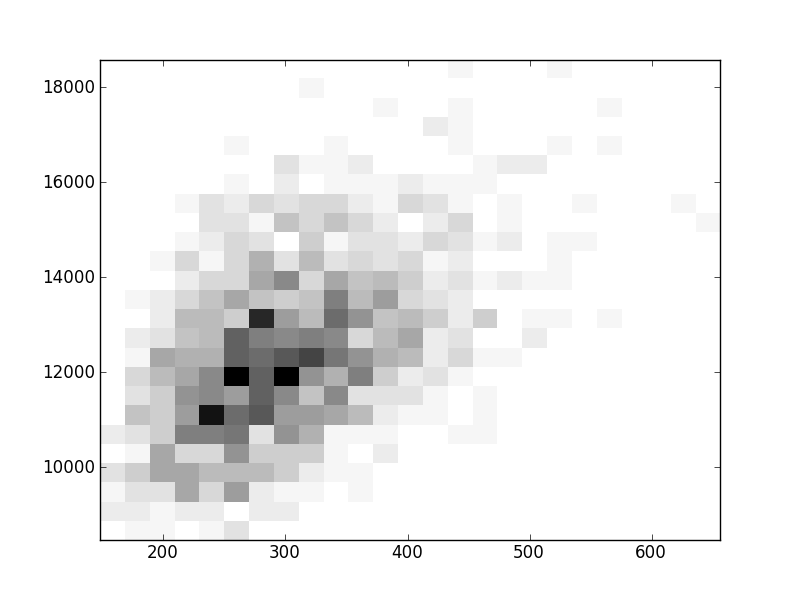 | 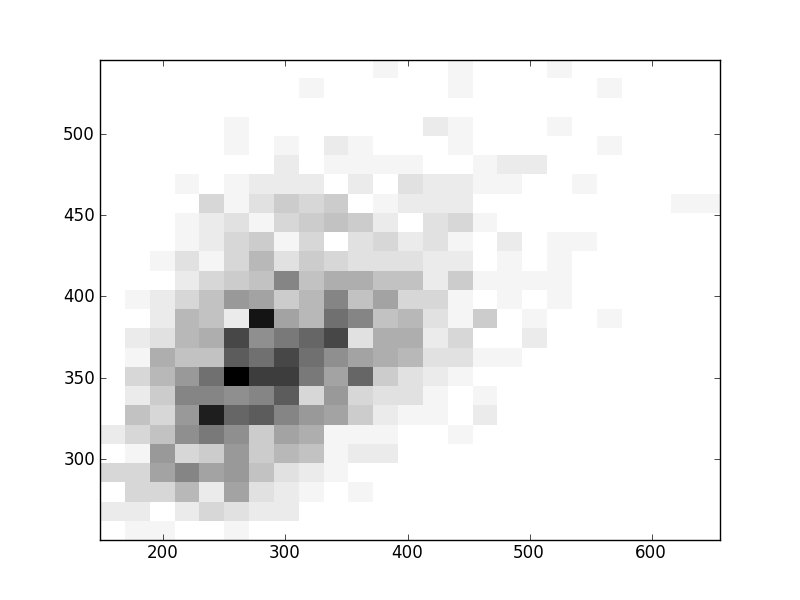 | 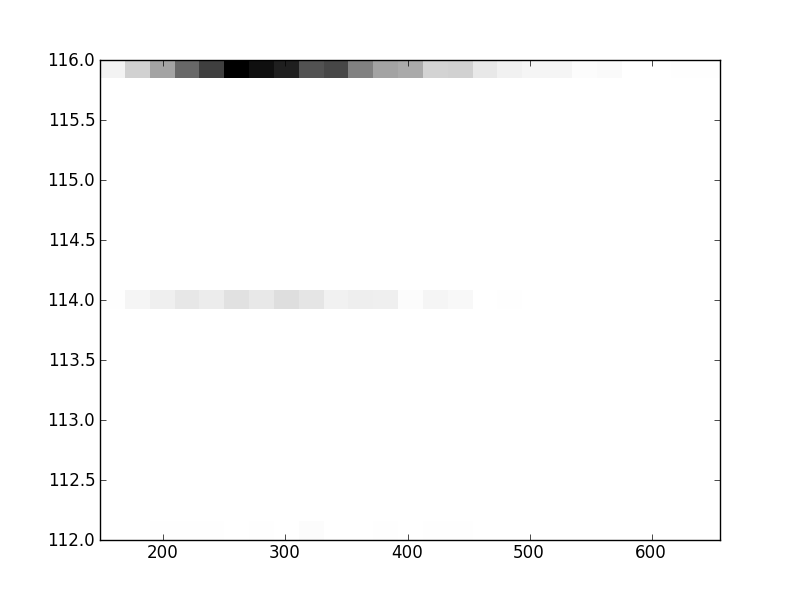 |
| Nsp p=1 | | |  | 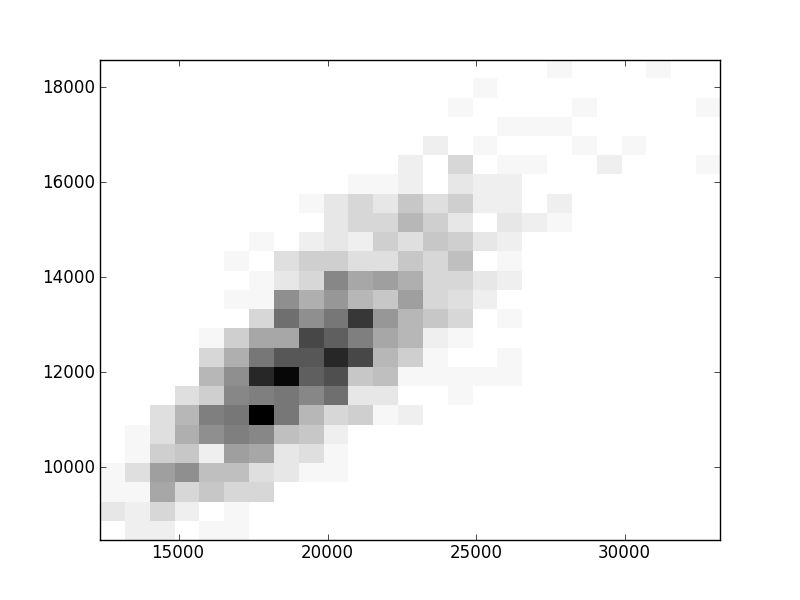 | 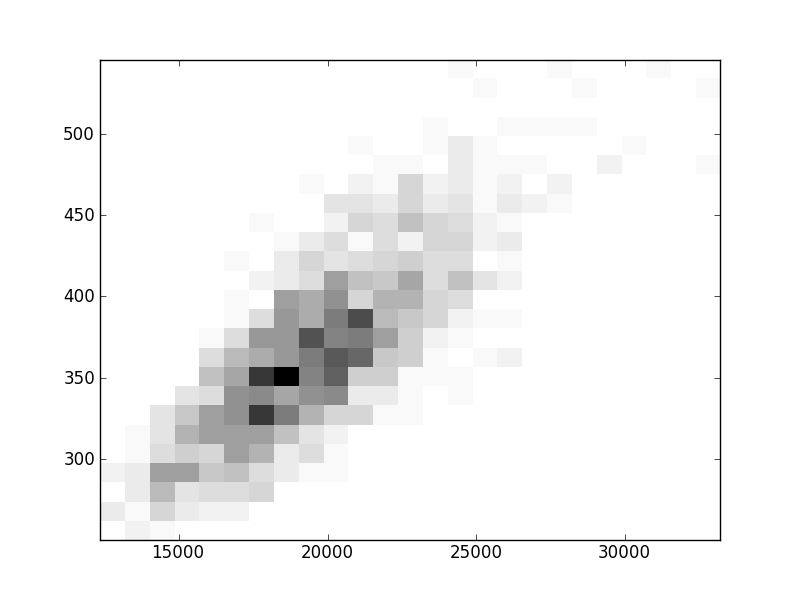 | 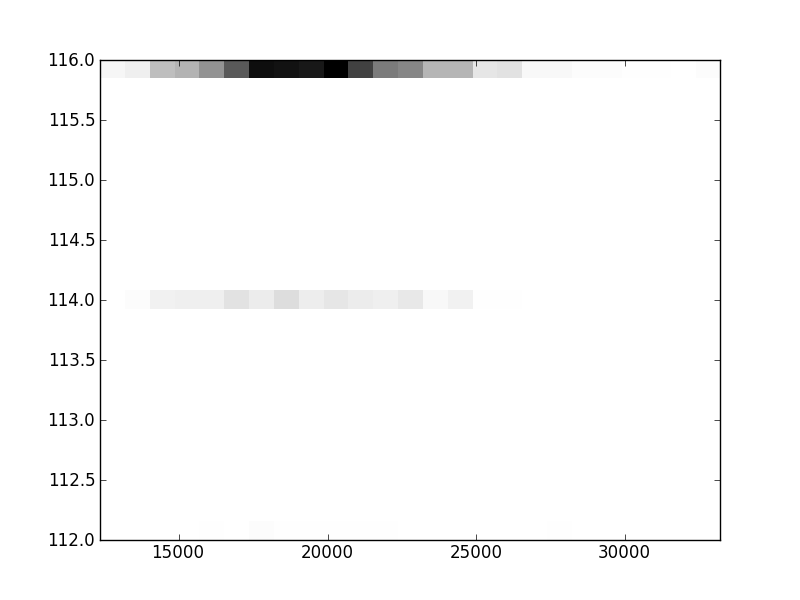 |
| Nsp p=2 | | | | 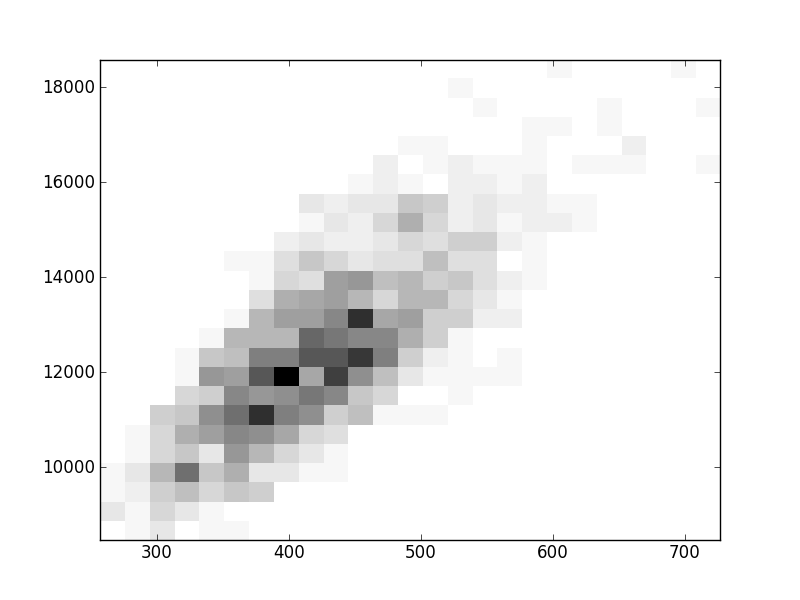 | 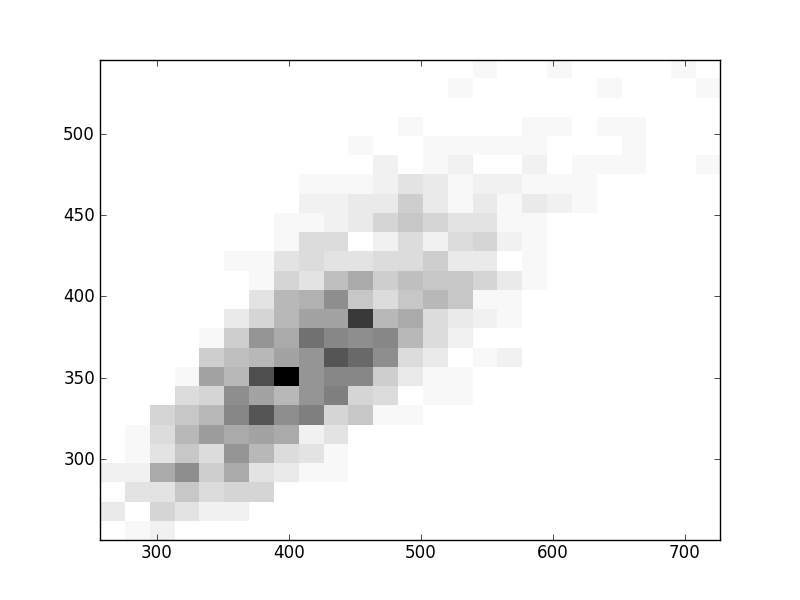 | 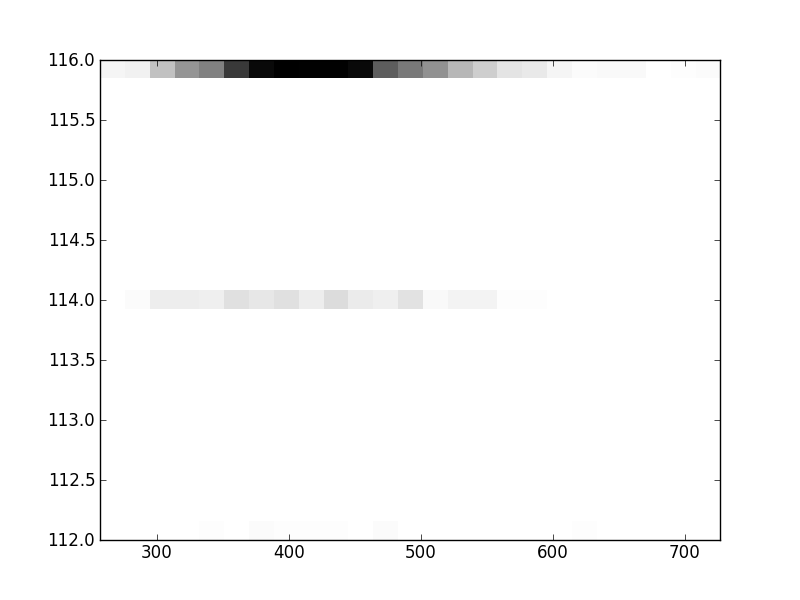 |
| Nunsp p=1 | | | | | 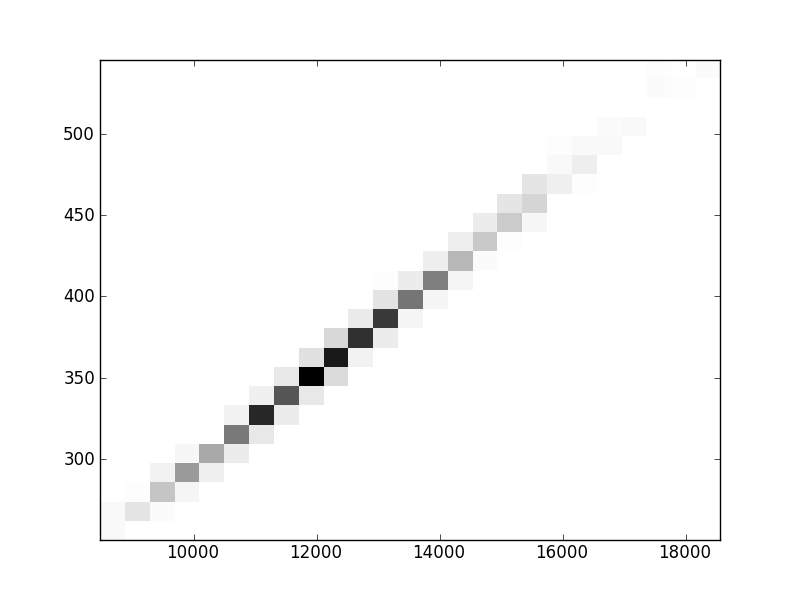 |  |
| Nunsp p=2 | | | | | |  |









