Cophenetic metrics for phylogenetic trees, after Sokal and Rohlf
Correlation between metrics over phylogenetic trees with 70 leaves (Simulation)
Non-necessary binary phylogenetic trees
Spearman's correlation between cophenetic, nodal and RF metrics.
| | Cophenetic p=1 | Cophenetic p=2 | Nodalsp p=1 | Nodalsp p=2 | RF |
| Coph p=1 | 1.0 | 0.985417388536 | 0.739183512282 | 0.76118357766 | 0.13699142899 |
| Coph p=2 | | 1.0 | 0.788629760704 | 0.811811901118 | 0.136486331967 |
| Nsp p=1 | | | 1.0 | 0.989350420264 | 0.202934397473 |
| Nsp p=2 | | | | 1.0 | 0.185446580154 |
| RF | | | | | 1.0 |
2D histograms. A darker squeare means that there are more trees with that relationship.
| | Cophenetic p=2 | Nodalsp p=1 | Nodalsp p=2 | RF |
| Coph p=1 |  | 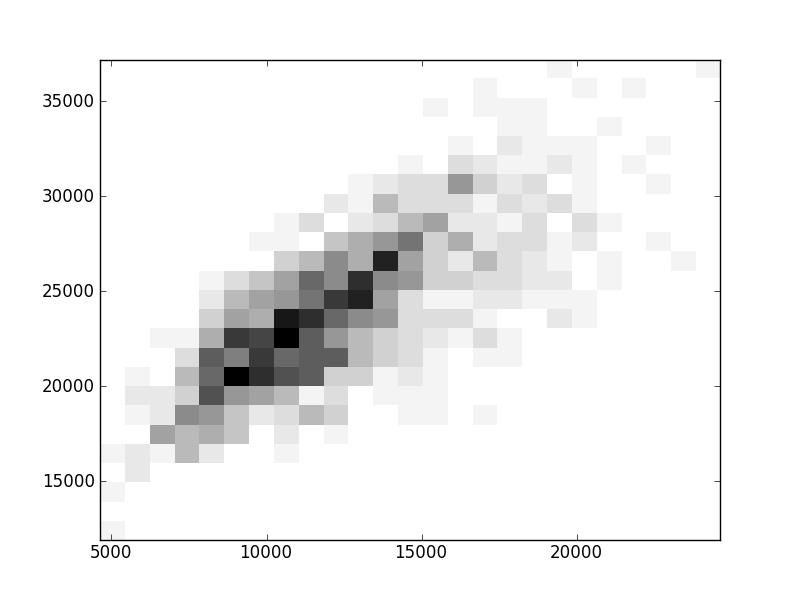 |  |  |
| Coph p=2 | |  |  |  |
| Nsp p=1 | | |  |  |
| Nsp p=2 | | | |  |
Binary phylogenetic trees
Spearman's correlation between cophenetic, nodal and RF metrics.
| | Cophenetic p=1 | Cophenetic p=2 | Nodalsp p=1 | Nodalsp p=2 | Nodalunsp p=1 | Nodalunsp p=2 | RF |
| Coph p=1 | 1.0 | 0.986141713808 | 0.742437102541 | 0.766324687494 | 0.433496636254 | 0.434450330025 | 0.0312820974475 |
| Coph p=2 | | 1.0 | 0.793372526483 | 0.819111267446 | 0.507654586302 | 0.509318965387 | 0.0321568711925 |
| Nsp p=1 | | | 1.0 | 0.990545013486 | 0.809959374746 | 0.809034046117 | 0.0480324771373 |
| Nsp p=2 | | | | 1.0 | 0.783696152513 | 0.785328264634 | 0.0437818679928 |
| Nunsp p=1 | | | | | 1.0 | 0.998033304132 | 0.0430765913515 |
| Nunsp p=2 | | | | | | 1.0 | 0.0401600497608 |
| RF | | | | | | | 1.0 |
2D histograms. A darker squeare means that there are more trees with that relationship.
| | Cophenetic p=2 | Nodalsp p=1 | Nodalsp p=2 | Nodalunsp p=1 | Nodalunsp p=2 | RF |
| Coph p=1 |  |  |  |  |  |  |
| Coph p=2 | |  |  |  |  |  |
| Nsp p=1 | | |  |  |  |  |
| Nsp p=2 | | | |  |  |  |
| Nunsp p=1 | | | | |  |  |
| Nunsp p=2 | | | | | |  |









