Cophenetic metrics for phylogenetic trees, after Sokal and Rohlf
Correlation between metrics over phylogenetic trees with 80 leaves (Simulation)
Non-necessary binary phylogenetic trees
Spearman's correlation between cophenetic, nodal and RF metrics.
| | Cophenetic p=1 | Cophenetic p=2 | Nodalsp p=1 | Nodalsp p=2 | RF |
| Coph p=1 | 1.0 | 0.98535328382 | 0.718200523684 | 0.744625060245 | 0.0887099096774 |
| Coph p=2 | | 1.0 | 0.772612385108 | 0.80161292207 | 0.0953695156285 |
| Nsp p=1 | | | 1.0 | 0.990013651429 | 0.210387992113 |
| Nsp p=2 | | | | 1.0 | 0.195582543952 |
| RF | | | | | 1.0 |
2D histograms. A darker squeare means that there are more trees with that relationship.
| | Cophenetic p=2 | Nodalsp p=1 | Nodalsp p=2 | RF |
| Coph p=1 |  |  |  |  |
| Coph p=2 | |  | 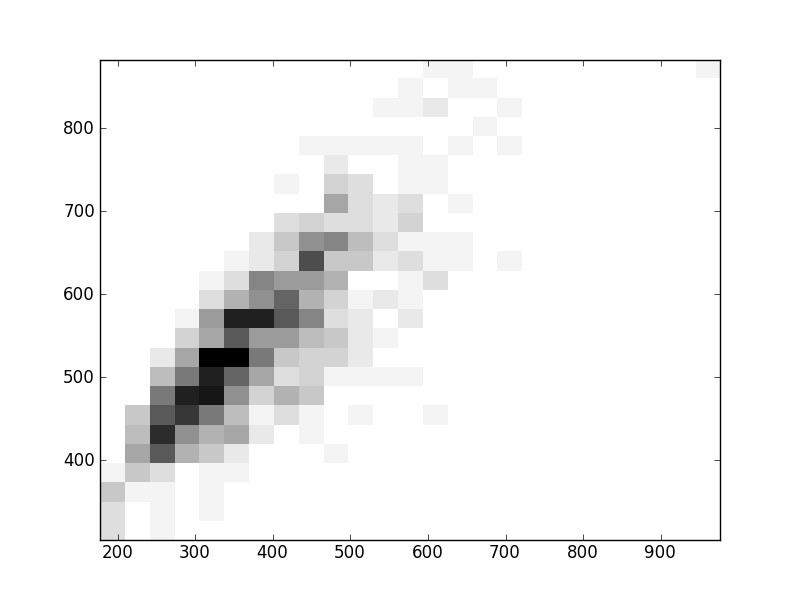 | 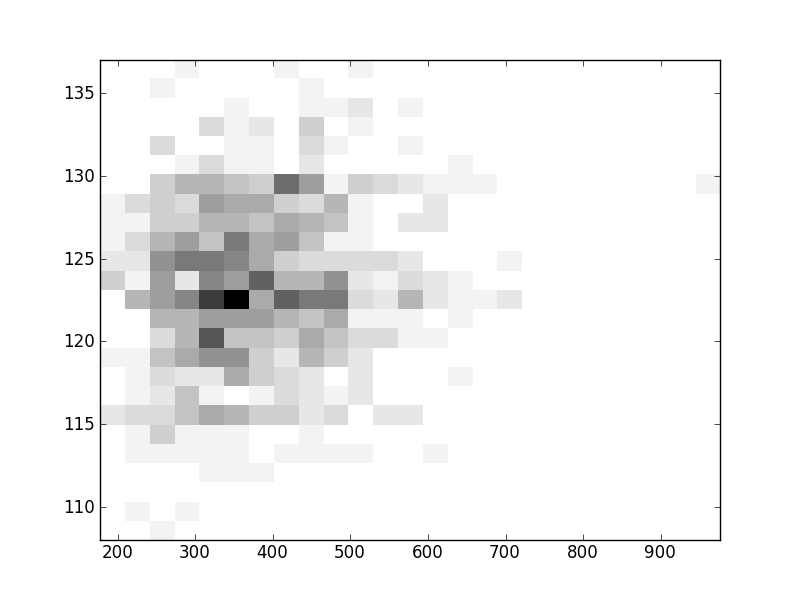 |
| Nsp p=1 | | |  |  |
| Nsp p=2 | | | |  |
Binary phylogenetic trees
Spearman's correlation between cophenetic, nodal and RF metrics.
| | Cophenetic p=1 | Cophenetic p=2 | Nodalsp p=1 | Nodalsp p=2 | Nodalunsp p=1 | Nodalunsp p=2 | RF |
| Coph p=1 | 1.0 | 0.987277836987 | 0.740944413224 | 0.763679271926 | 0.427632264403 | 0.428086160429 | -0.0200678869699 |
| Coph p=2 | | 1.0 | 0.789148509281 | 0.813702343988 | 0.49357920962 | 0.494296100975 | -0.0201830151231 |
| Nsp p=1 | | | 1.0 | 0.98971928129 | 0.805166431852 | 0.803240074358 | -0.00569733747464 |
| Nsp p=2 | | | | 1.0 | 0.778205786461 | 0.779079706026 | -0.00988479210178 |
| Nunsp p=1 | | | | | 1.0 | 0.99808899562 | 0.0439895828445 |
| Nunsp p=2 | | | | | | 1.0 | 0.0453588968198 |
| RF | | | | | | | 1.0 |
2D histograms. A darker squeare means that there are more trees with that relationship.
| | Cophenetic p=2 | Nodalsp p=1 | Nodalsp p=2 | Nodalunsp p=1 | Nodalunsp p=2 | RF |
| Coph p=1 |  |  |  |  |  | 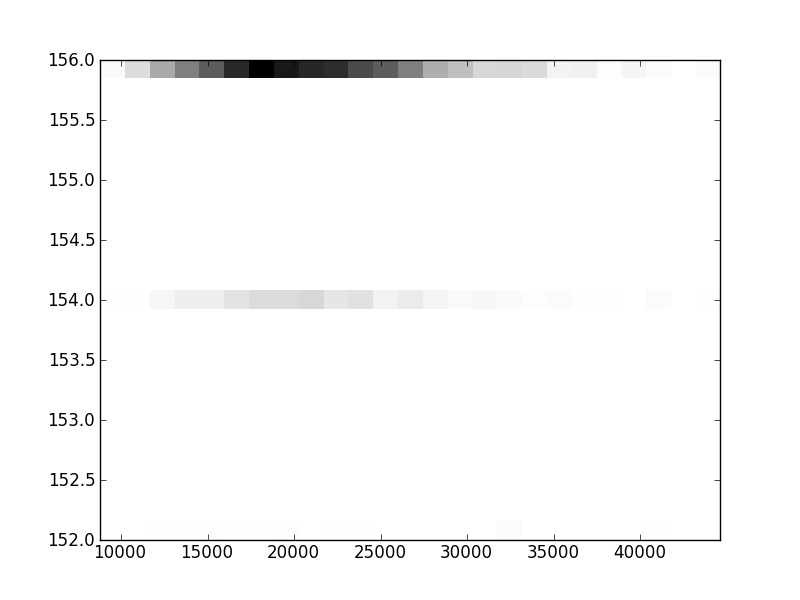 |
| Coph p=2 | |  |  |  |  | 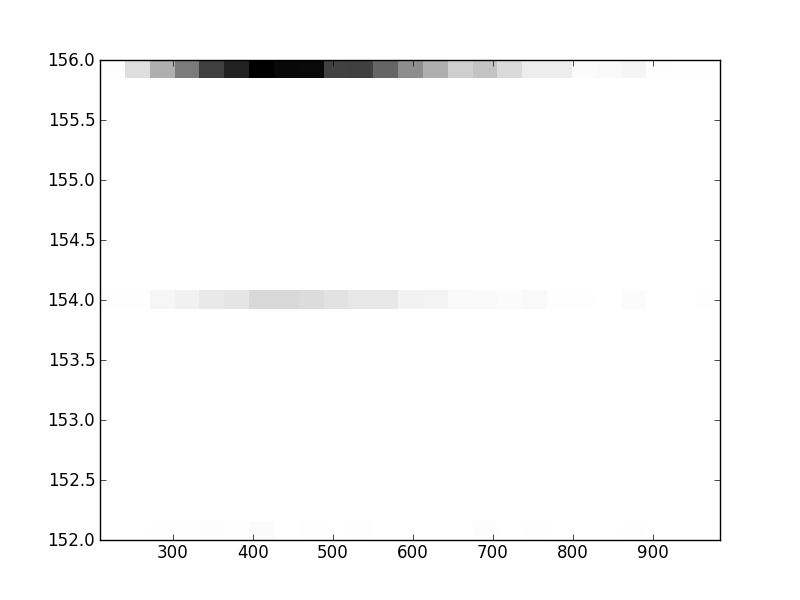 |
| Nsp p=1 | | |  |  |  |  |
| Nsp p=2 | | | |  |  |  |
| Nunsp p=1 | | | | | 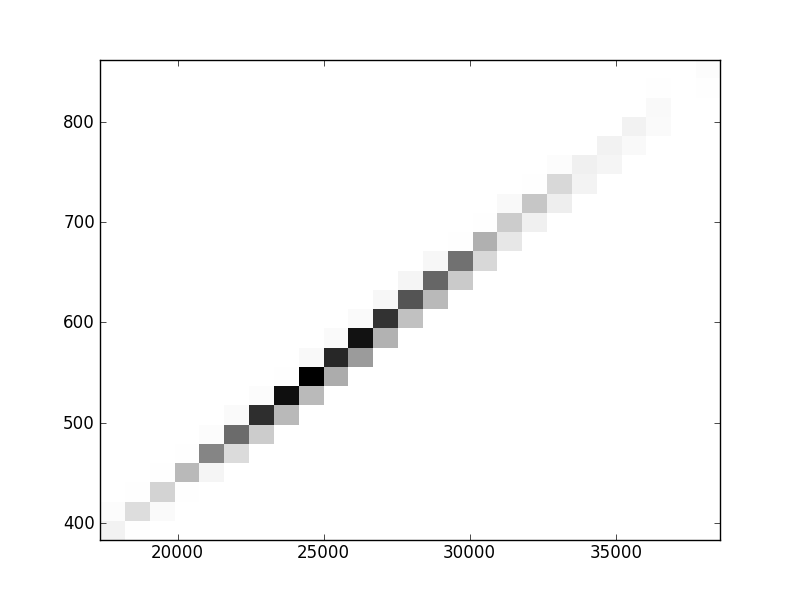 |  |
| Nunsp p=2 | | | | | |  |









