Cophenetic metrics for phylogenetic trees, after Sokal and Rohlf
Correlation between metrics over phylogenetic trees with 90 leaves (Simulation)
Non-necessary binary phylogenetic trees
Spearman's correlation between cophenetic, nodal and RF metrics.
| | Cophenetic p=1 | Cophenetic p=2 | Nodalsp p=1 | Nodalsp p=2 | RF |
| Coph p=1 | 1.0 | 0.986503849065 | 0.712786952902 | 0.738727339518 | 0.154958461926 |
| Coph p=2 | | 1.0 | 0.768963674346 | 0.795044586125 | 0.160599582492 |
| Nsp p=1 | | | 1.0 | 0.991165696545 | 0.232420905806 |
| Nsp p=2 | | | | 1.0 | 0.215037407596 |
| RF | | | | | 1.0 |
2D histograms. A darker squeare means that there are more trees with that relationship.
| | Cophenetic p=2 | Nodalsp p=1 | Nodalsp p=2 | RF |
| Coph p=1 |  | 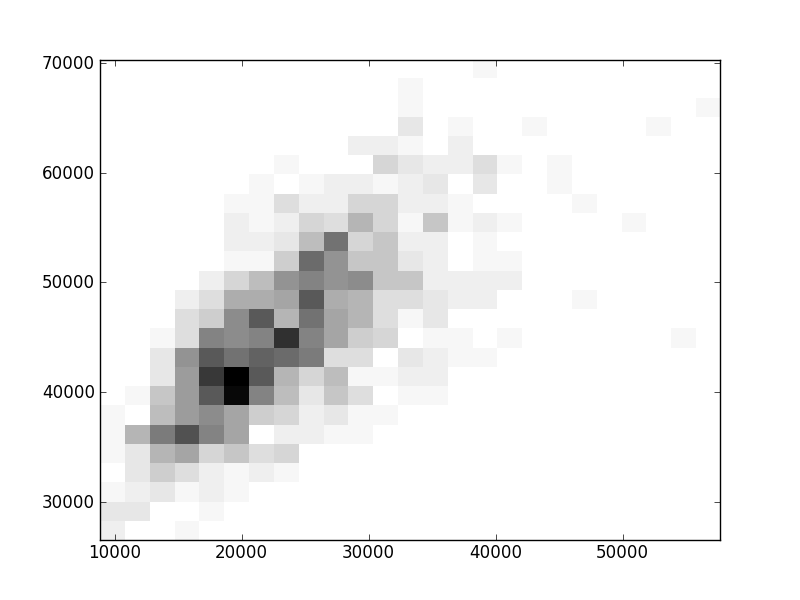 | 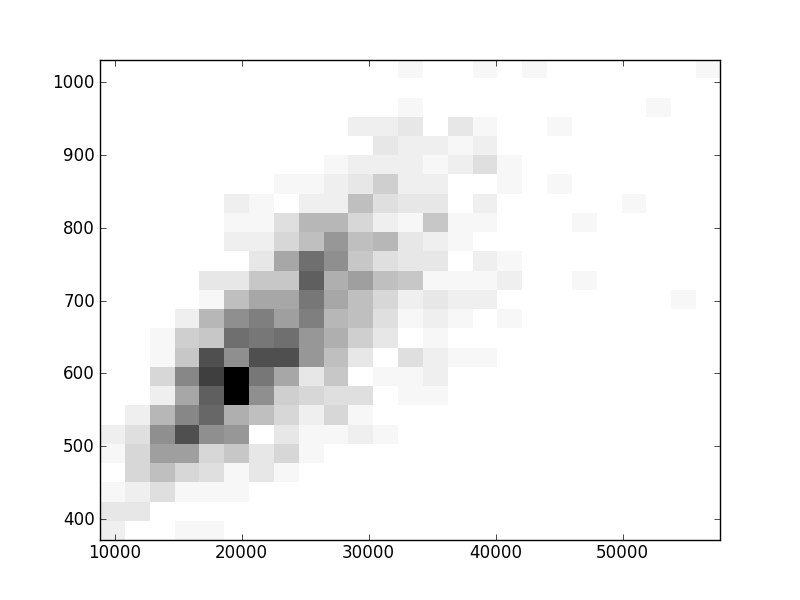 |  |
| Coph p=2 | |  |  |  |
| Nsp p=1 | | |  |  |
| Nsp p=2 | | | |  |
Binary phylogenetic trees
Spearman's correlation between cophenetic, nodal and RF metrics.
| | Cophenetic p=1 | Cophenetic p=2 | Nodalsp p=1 | Nodalsp p=2 | Nodalunsp p=1 | Nodalunsp p=2 | RF |
| Coph p=1 | 1.0 | 0.986502171703 | 0.728397007089 | 0.751964423605 | 0.41934282485 | 0.418718440941 | -0.0243802449481 |
| Coph p=2 | | 1.0 | 0.777203446652 | 0.802738057332 | 0.487025554474 | 0.486733263417 | -0.0228476623853 |
| Nsp p=1 | | | 1.0 | 0.990327675596 | 0.810140151857 | 0.808487162944 | 0.00235941507282 |
| Nsp p=2 | | | | 1.0 | 0.781095933558 | 0.781873895861 | 0.00454238350818 |
| Nunsp p=1 | | | | | 1.0 | 0.998167606475 | 0.0221248949417 |
| Nunsp p=2 | | | | | | 1.0 | 0.0222513405397 |
| RF | | | | | | | 1.0 |
2D histograms. A darker squeare means that there are more trees with that relationship.
| | Cophenetic p=2 | Nodalsp p=1 | Nodalsp p=2 | Nodalunsp p=1 | Nodalunsp p=2 | RF |
| Coph p=1 |  |  |  |  |  |  |
| Coph p=2 | | 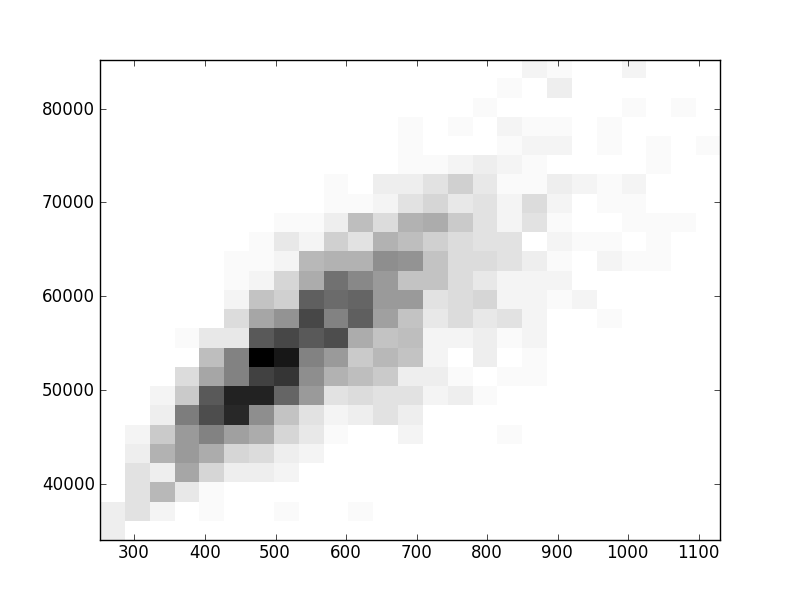 |  |  | 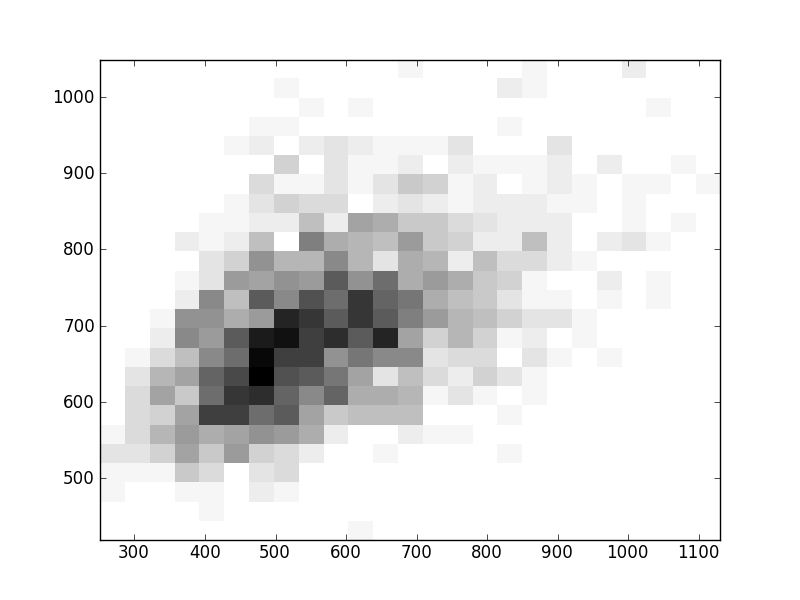 |  |
| Nsp p=1 | | |  | 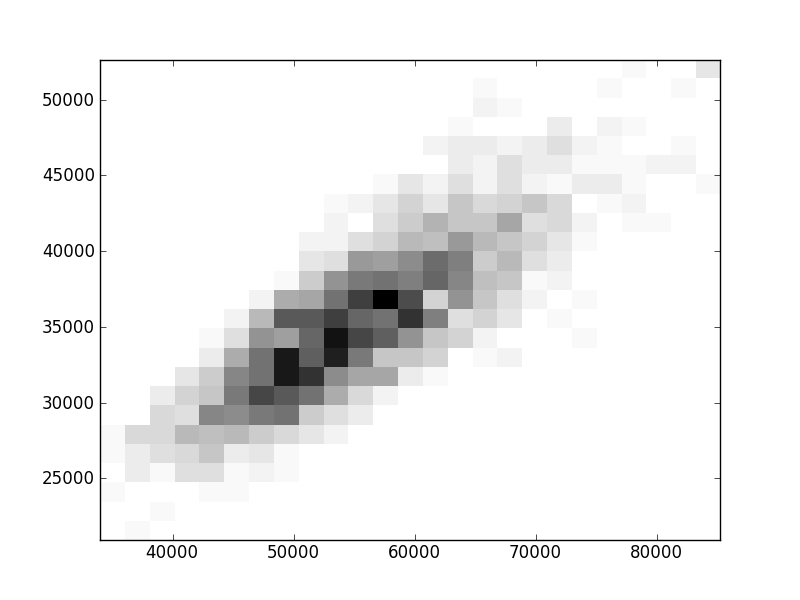 | 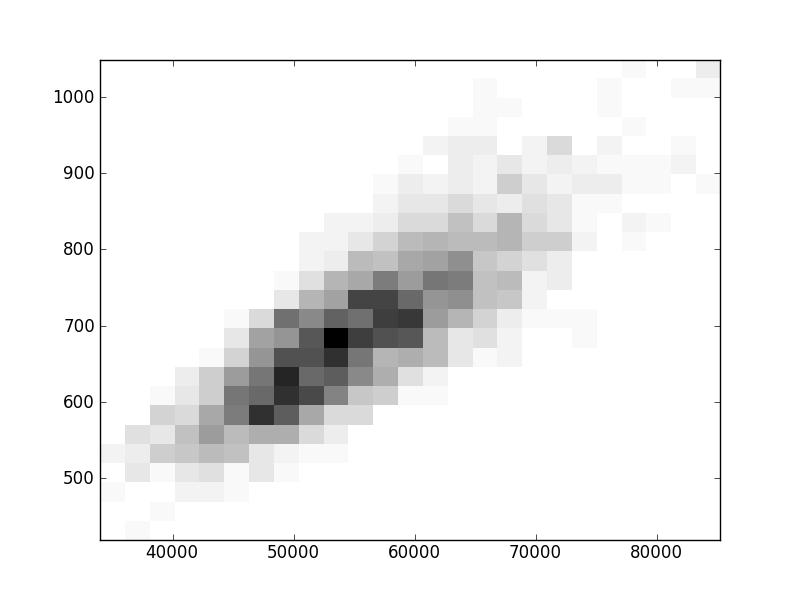 |  |
| Nsp p=2 | | | |  |  |  |
| Nunsp p=1 | | | | | 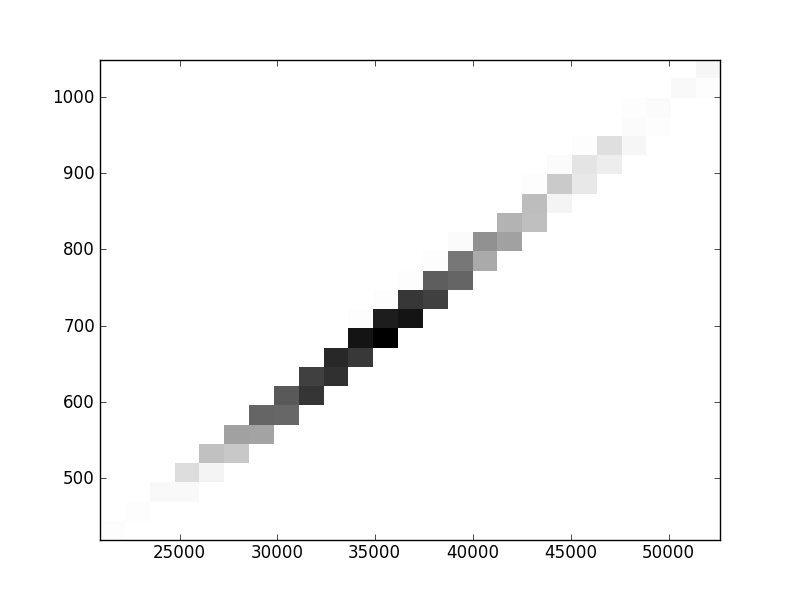 |  |
| Nunsp p=2 | | | | | |  |









